
Engineering Specificity into Unspecific Peroxygenases (UPOs)
March 3, 2025
Pushing Boundaries: Biocatalysis Surpassing Conventional Chemistry in Unprecedented Ways
Biocatalysis
Engineering Specificity into Versatile Peroxygenases for Industrial Biocatalysis
Peroxygenases are a fascinating class of enzymes that are gaining a lot of traction in biocatalysis due to their unique and versatile catalytic capabilities. They are capable of performing a wide array of oxyfunctionalizations of non-activated carbons.
This enzyme class is mechanistically simpler than P450BM3 as it requires only hydrogen peroxide as the terminal oxidant without the need for cofactors and appropriate cofactor recycling systems. Making them easier to use in an industrial manufacturing setting.
However, unspecific peroxygenases (UPOs) have been shown to exhibit remarkably high substrate promiscuity. Our goal here was to begin with a wildtype (WT) enzyme that could catalyze many different reactions and accept many different substrates, then use ultrahigh throughput screening (uHTS) to engineer specificity into the biocatalyst (Fig. 1).
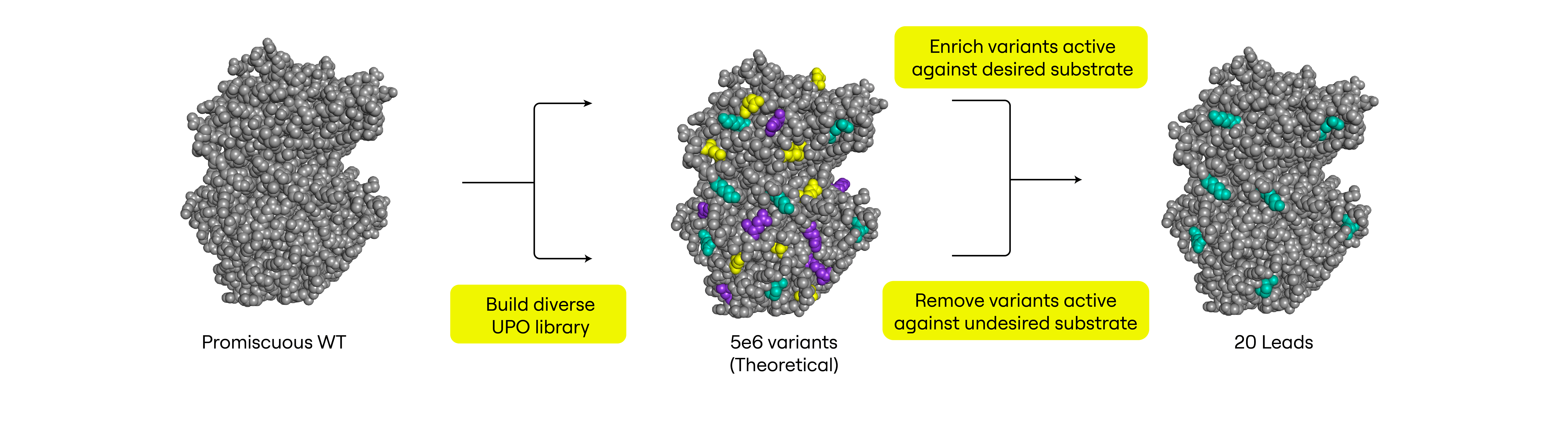
Building a Diverse Library of AbrUPO Variants in Pichia pastoris
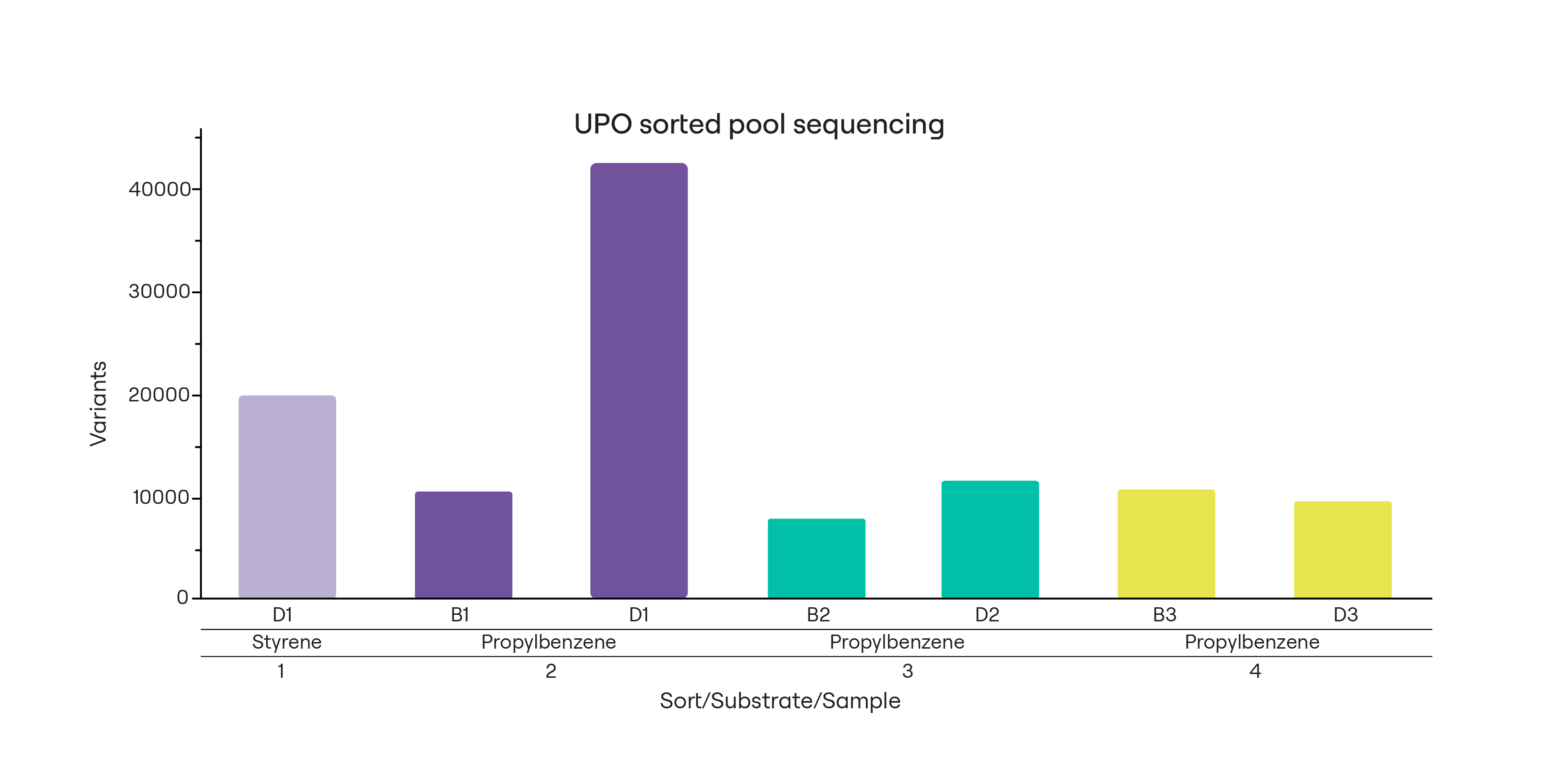
For this work, we built a diverse library of AbrUPO variants, previously shown to catalyze production of at least 51 products from 16 different substrates.1 We expressed this library in Pichia pastoris, chosen because it can perform glycosylation and it is known to secrete heterologous proteins. The theoretical diversity of this library was 5 x 106 variants, and long-read sequencing confirmed at least 104-105 variants in the library (Fig. 2).
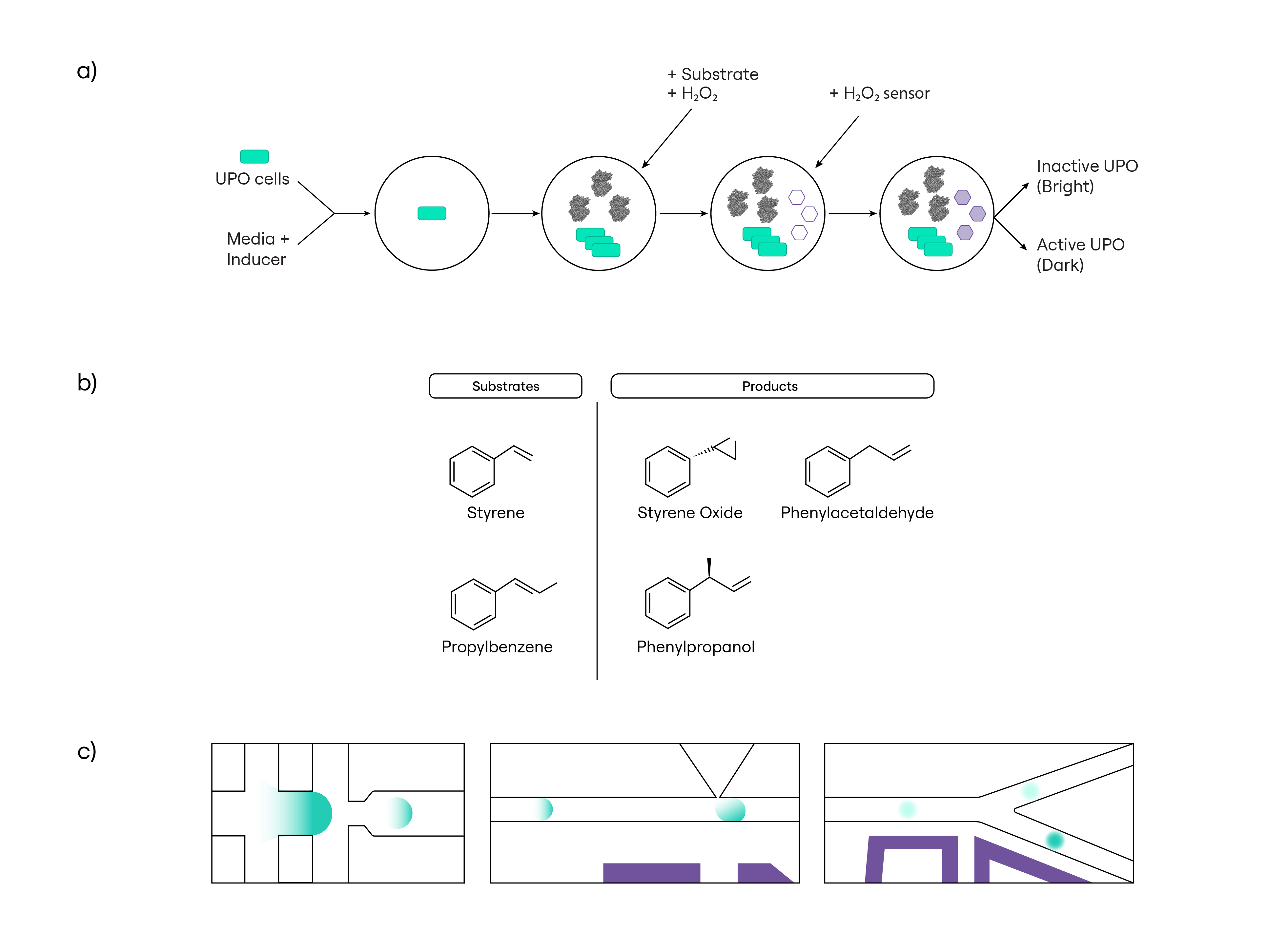
Screening UPO Activity Using a Fluorimetric Microfluidics Assay
We developed a fluorimetric assay compatible with our microfluidics platform that allowed us to screen this library for variants that were active against either a desired substrate, styrene, or an undesired substrate, propylbenzene (Fig. 3).
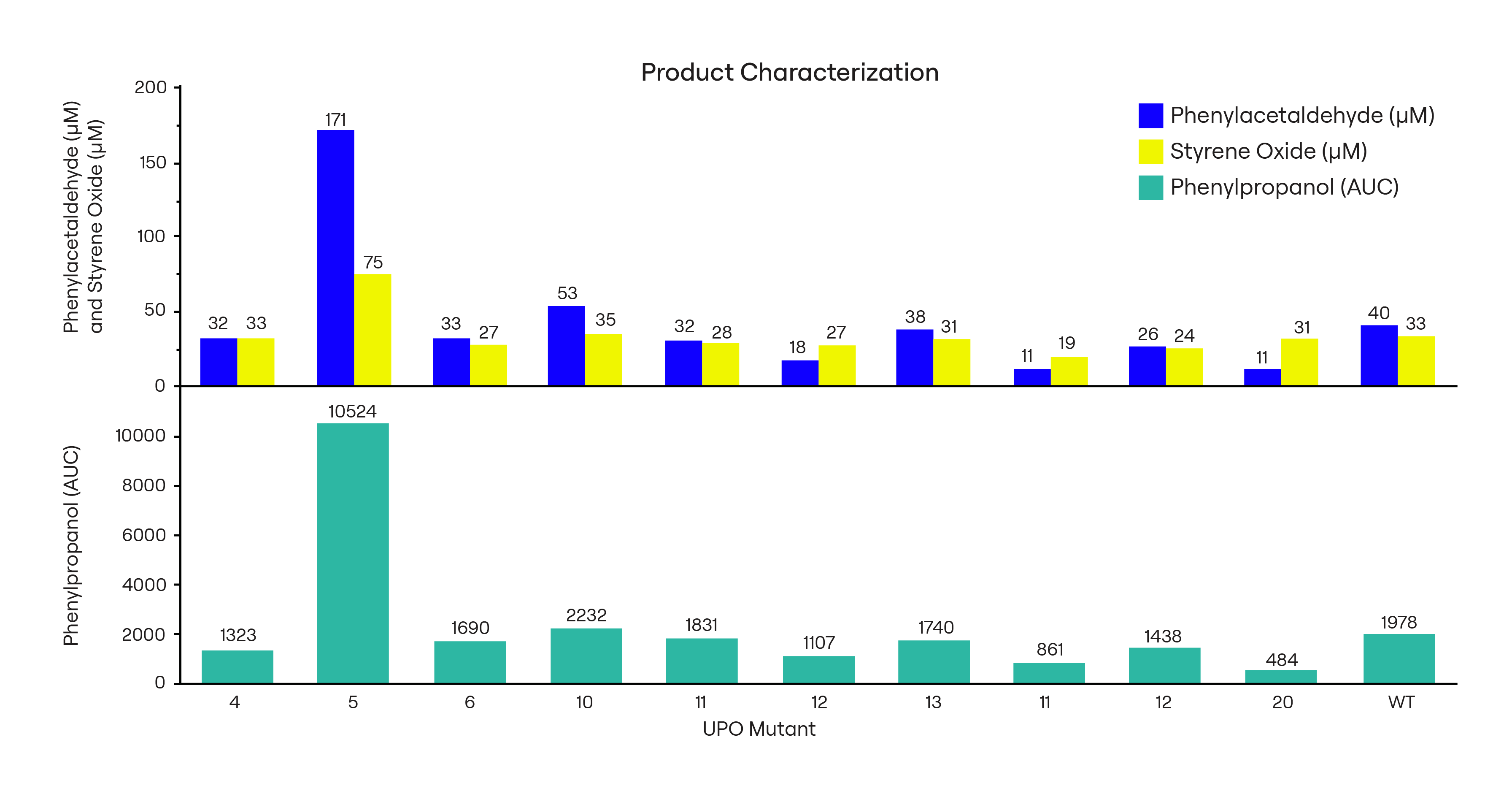
Validating UPO Variants with GC and HPLC Analysis
Of the tested lead compounds, UPO 5 is more active than WT, producing 3-5x more of each product, and UPO 20 is more specific than WT, producing 78% as much styrene oxide, 33% as much phenylacetaldehyde, and 24% as much phenylpropanol as WT. (Fig. 4).
Conclusion
In a single round of directed evolution, we screened a diverse library containing at least 104 AbrUPO variants expressed in Pichia pastoris. Using a modular microfluidics assay, we engineered one AbrUPO variant that produces purer styrene oxide product and another variant that catalyzes 3-5x more production than the parent enzyme.
1Schmitz, F. et. al., React. Chem. Eng., 2023.